Whole plasmid sequencing

Get the whole picture with fast results and detailed reports. Submit purified DNA, or skip the minipreps with our PrepLESS Flow.
Data and deliverables
- Consensus sequence (.fasta, .gbk)
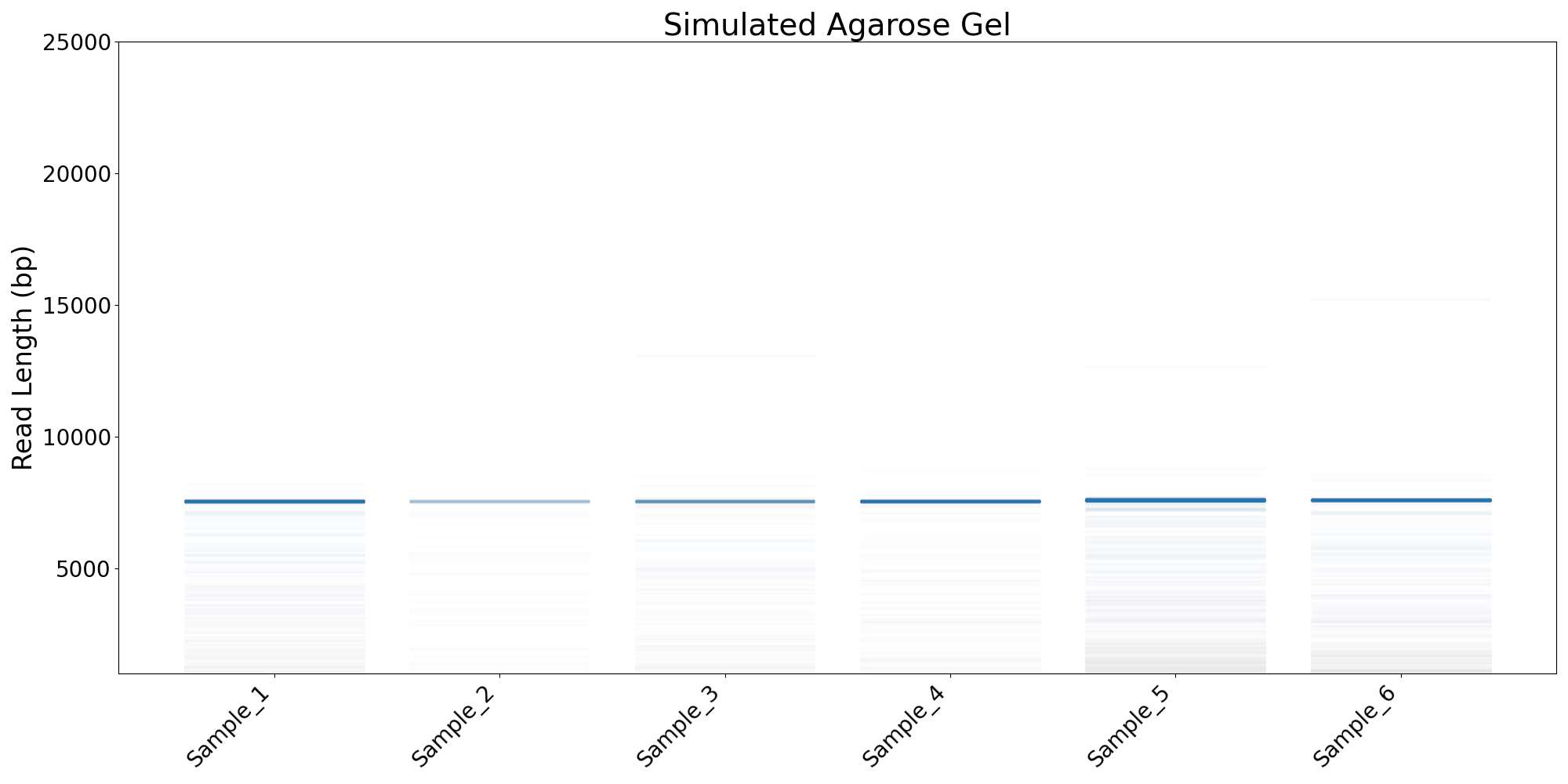
- Simulated agarose gel (.png)
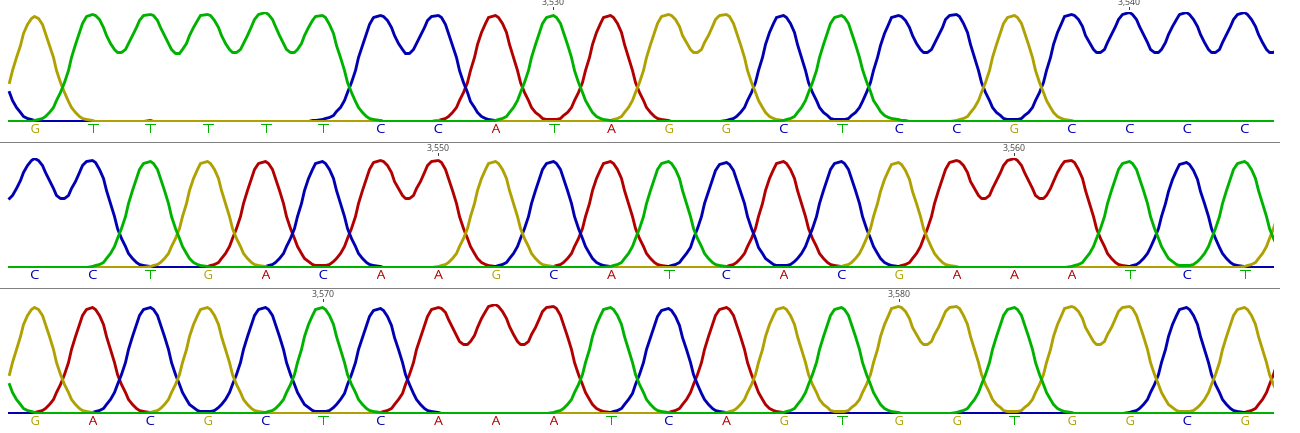
- Chromatogram (.ab1)
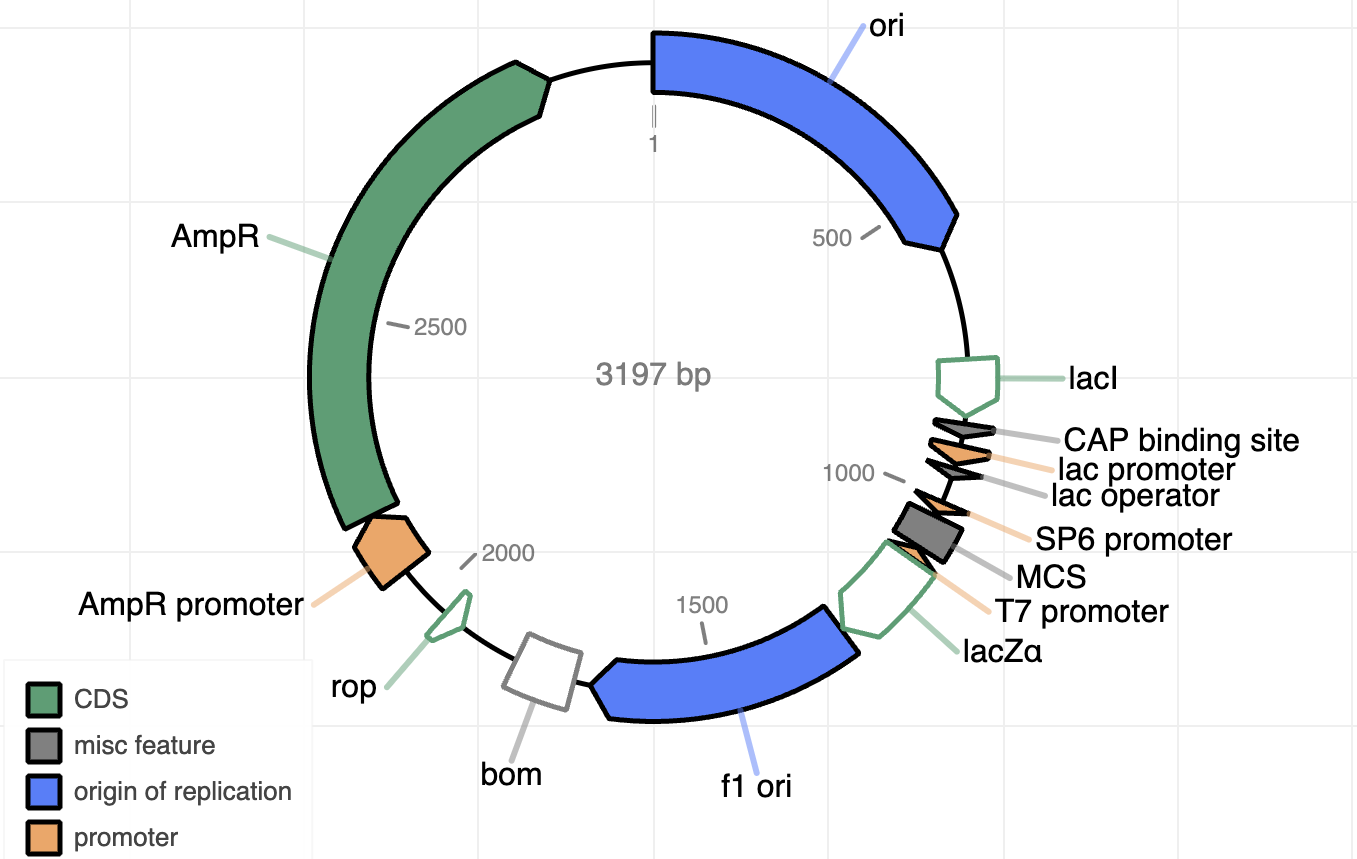
- Annotated map (.html, .gbk)
- Data quality report (.html)
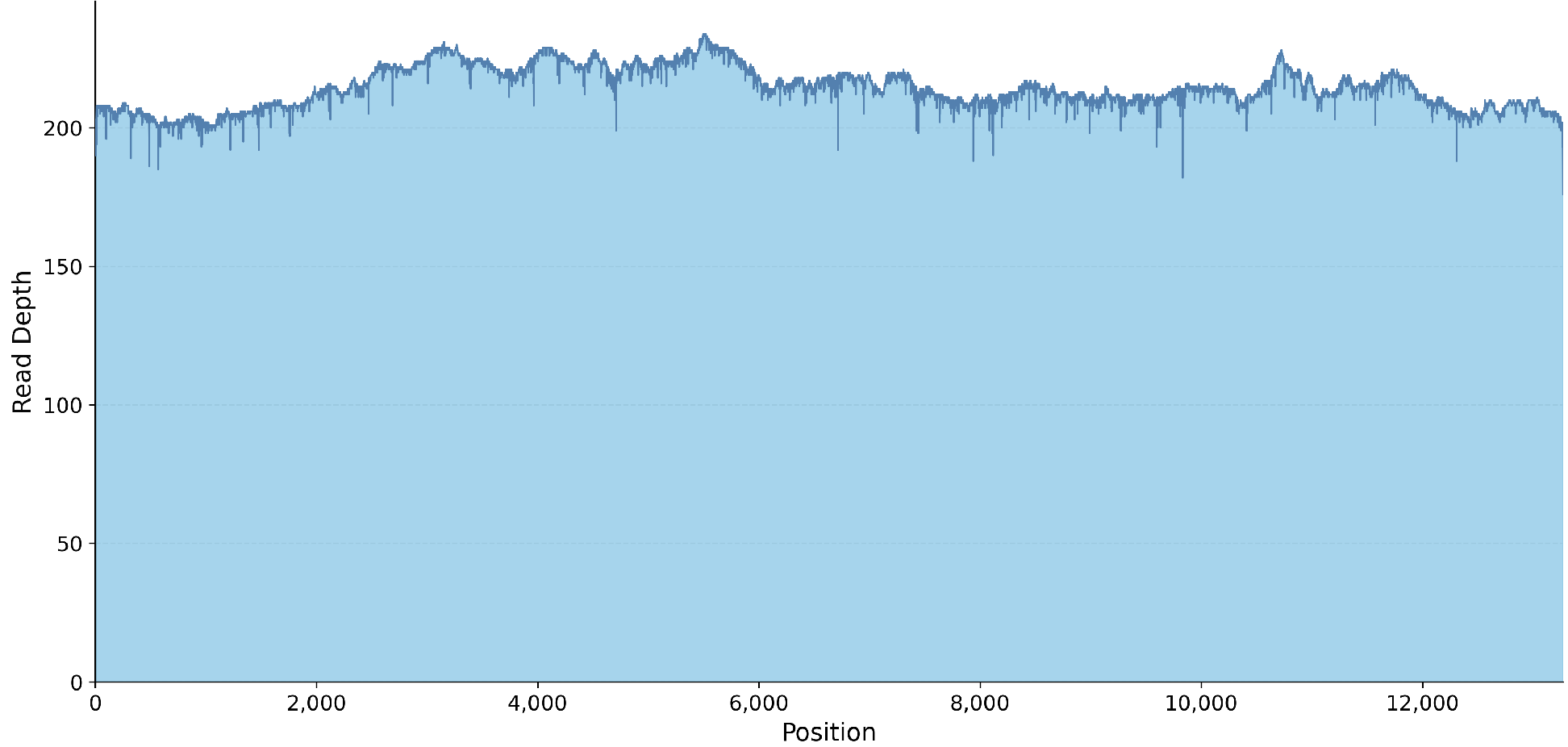
- Coverage plot (.png)
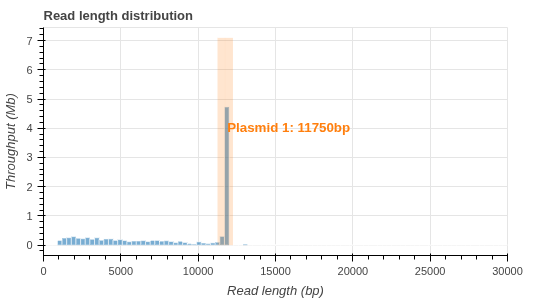
- Read length histogram (.png)
- Raw sequencing reads (.fastq)
- Per-base sequencing information (.txt)
- Responsive customer support
Pricing
All prices shown are $CAD and subject to local sales tax.
Purified plasmids
PrepLESS Flow
Claim back your time- we'll extract your plasmids for you.
FAQ
How accurate are my sequencing results?
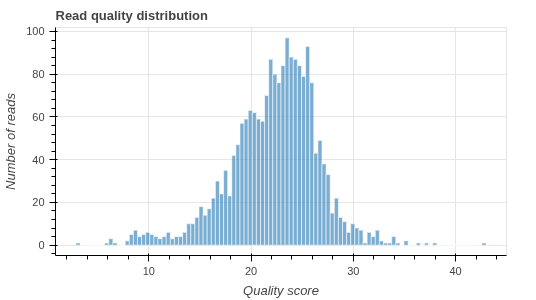
Consensus sequences we return typically have accuracy scores above Q50, or 1 error in 100 000 bases. For plasmids and amplicons without homopolymers, this typically means perfection throughout the whole plasmid.
We use the latest generation of Oxford Nanopore Technologies long-read sequencing platform, with their best basecalling models. The remaining errors are often due to modified bases or homopolymer stretches.
How much data do I get?
For several services, we provide data targets when DNA meets our input quality requirement. Since throughput is directly related to the quality and size of input DNA, samples that don't meet our concentration or purity requirements may not hit the data target.
For services without a data target, we do not provide a guaranteed number of reads or bases. As long as ~20X coverage is obtained, we consider sequencing a success since this consensus will be high quality.
What quality and concentration is required?
Since we use a PCR-free method for sequencing, sufficient input mass and pure DNA is essential for success. DNA concentration requirements may vary by service type.
Our standard plasmid requires a minimum concentration anywhere between 20 - 200 ng / µL, measured by Qubit (Nanodrop often over-estimates concentration, especially at lower concentrations). Pure DNA measured by Nanodrop typically has an A260/280 of 1.8 and a A260/230 of 2.0-2.2. The most common cause of sequencing failure is when there is insufficient DNA or DNA is contaminated (e.g., RNA contamination).
What is your re-sequencing policy if a sample fails?
While our protocols are robust for the vast majority of samples that meet our input requirements, sequencing failure can occur when DNA does not meet our input requirements, or contamination is present.
Our team will evaluate if re-sequencing is likely to produce a success results - re-sequencing of the submitted sample is performed free of charge when it's likely to produce a successful result. Depending on the root cause, we may be able modify our protocols to improve coverage. An example where re-sequencing will likely not be successful is when sufficient throughput is collected, but the vast majority of the bases collected is found to be gDNA contamination.
We still charge for failed samples since they consume reagents.